WHO experts estimated that the annual number of deaths caused by antibiotic resistance will increase from the current 700,000 to 10 million by 2050, which could cost the global economy 100 trillion dollars. One major cause behind the ever-increasing emergence and spread of antibiotic-resistant bacteria is the misuse and overuse of antibiotics in treating bacteria-infected patients. At present, the conventional phenotypic antibiotic susceptibility test (AST) for pathogenic bacteria (as shown in Figure 1) typically takes 3-7 days to obtain analysis results of pathogen identification and antibiotic sensitivity of patient samples. To determine an efficacious antibiotic for an infecting pathogen and reduce the need for broad-spectrum antibiotics, it is vital to rapidly identify the antibiotic susceptibility profile of the pathogen.
Recently, the SONG Yizhi research team from Suzhou Institute of Biomedical Engineering and Technology of Chinese Academy of Sciences, the WANG Minggui research team from Huashan Hospital affiliated to Fudan University, in collaboration with the HUANG Wei research team from the University of Oxford developed a fast Raman-assisted antibiotic susceptibility test (FRAST) utilizing single cell Raman spectroscopy coupled with heavy water (D2O) labeling, which applies to blood and urine samples. Under optimal conditions, the sample-to-report time of FRAST could be reduced to 3 hours for urine samples and 21 hours for sepsis samples.
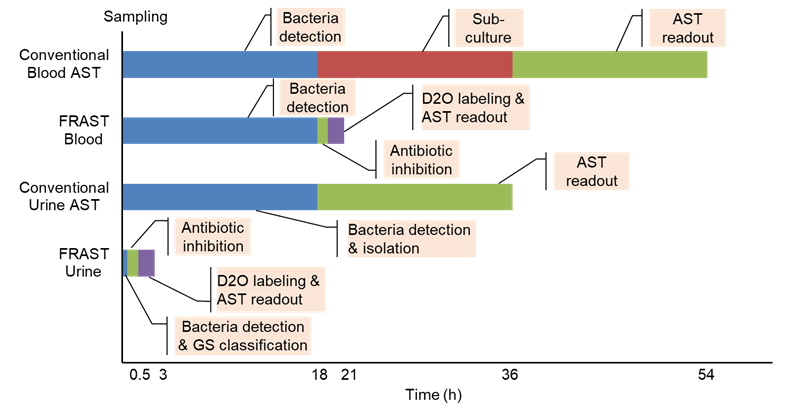
Figure 1. Turnaround time (TAT) required for implementing the conventional AST and FRAST for blood and urine samples.(Image by SIBET)
FRAST is based on the combination of Raman spectroscopy and D2O labeling, and its principle is that metabolically active bacteria in the presence of D2O will take in deuterium from D2O via nicotinamide adenine dinucleotide phosphate (NADPH) and add D into biomolecules to form carbon-deuterium (C-D) bonds. The cells containing C-D bonds show a distinguishable Raman band shifted from C-H vibration, and the band can be served as a distinct biomarker for the metabolic activity of a single cell. Under the effect of antibiotics, the susceptible bacteria do not show this band due to antibiotic inhibition of the metabolic activity.
The FRAST procedures are shown in Figure 2. For urinary samples, the bacteria were collected by centrifugation, and then were observed under a confocal Raman microscope to capture the Raman fingerprints, which can reflect the number of bacteria in the samples. After that, the collected fingerprints were matched with the database of Gram-negative and Gram-positive bacteria by machine learning models to identify whether the bacteria are Gram-positive or negative, so as to determine an appropriate susceptibility plate. Then, the urine samples were added to the plate and incubated for 1 hour before adding D2O. After being exposed to D2O for 1 hour, the samples were centrifuged and washed to collect Raman spectra. The minimum inhibitory concentration (MIC) was then calculated according to the intensity of C-D peak after applying antibiotics. For sepsis samples, they were cultivated in aerobic blood bottles in a blood cultivation system until it flagged positive, then the same method was adopted to collect Raman spectra and calculate the MIC value.
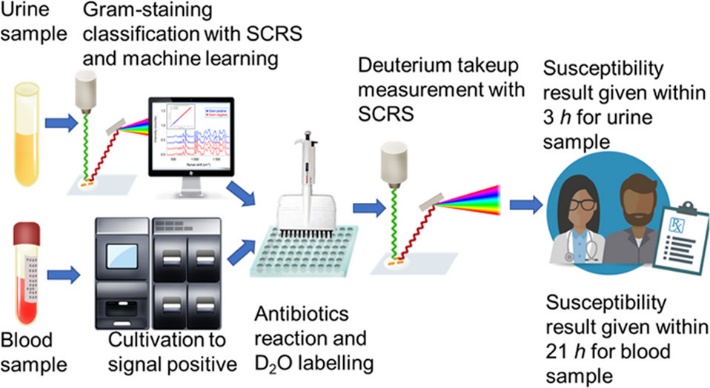
Figure 2. The flow chart of FRAST procedures for clinical urine and blood samples.(Image by SIBET)
This pilot work carried out around 3,200 experiments and acquired about 64,000 single-cell Raman spectra. A large amount of data has provided a comprehensive and large-scale evaluation of the Raman deuterium-labeling AST assay. As a result, the FRAST and the gold standard (the broth microdilution method or automated susceptibility testing system) showed high consistency. Compared with other Raman-DIP-based studies on pathogens’ susceptibility to antibiotics, this study proved for the first time that the combination of single cell Raman spectroscopy and D2O labeling can be used to analyze the drug resistance of pathogens in urine or blood samples. Besides, integrating with the Raman-based approach for Gram stain classification makes FRAST a relatively independent and complete test method. Therefore, clinicians can rapidly identify antibiotic susceptibility of pathogens without other assistance. Different from the molecular diagnosis of drug resistance developed in recent years, FRAST is based on the phenotype of the bacteria treated with antibiotics, and there will be no misjudgment on antibiotic susceptibility due to unknown drug resistance mechanisms or gene expression regulation.
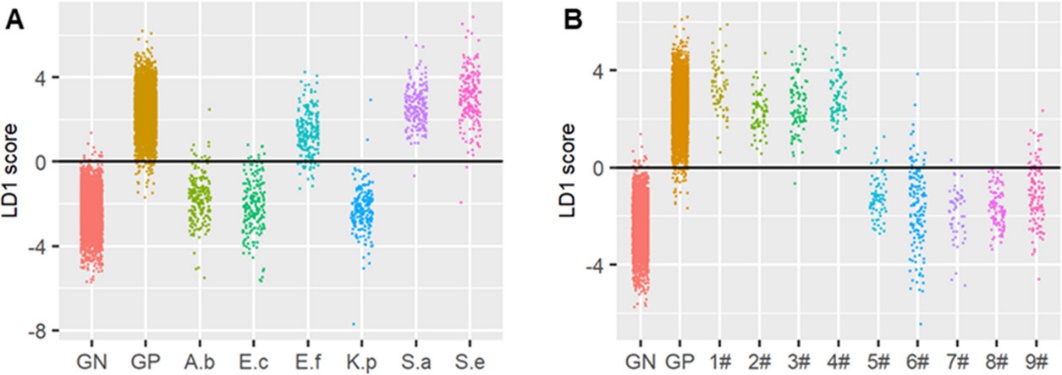
Figure 3. Gram stain classification of pathogenic bacteria was accurately predicted by FRAST.(Image by SIBET)
The research article “Development of a Fast Raman-Assisted Antibiotic Susceptibility Test (FRAST) for the Antibiotic Resistance Analysis of Clinical Urine and Blood Samples” has been published in Analytical Chemistry. This research is supported by the National Key R&D Program of China, Scientific Instrument Developing Project of the Chinese Academy of Sciences, etc.
Contact
XIAO Xintong
Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences (http://www.sibet.cas.cn/)
Phone: 86-512-69588013
E-mail: xiaoxt@sibet.ac.cn