Nucleic acids analysis is mainly applied on pathogen detection, genetic disease identification, early diagnosis of cancers. For example, quantitative analysis of circulating tumor DNA (ctDNA), a free DNA fragment derived from malignant cells which carries tumor specific sequence changes, can help obtain abundant information about tumors, including gene point mutation, genome integrity. Therefore, ctDNA is regarded as a personalized tumor marker, which plays a key role in cancer diagnosis and malignancy evaluation.
Recently, MIAO Peng’s group from Suzhou Institute of Biomedical Engineering and Technology (SIBET) developed an electrochemical DNA nanomachine based on looped bipedal DNA walking reaction for highly sensitive analysis of nucleic acids.
MIAO and his team built a pH controllable intermolecular triple helix DNA nanostructure between DNA probes A and B through sequence design, and then constructed a renewable modified electrode interface.
They designed a simple but effective strand displacement amplification strategy to amplify the information of target sequence. “Through the integration of primer and template into a single hairpin structured DNA probe, the reaction rate was effectively improved,” said MIAO.
In the presence of target sequence, a large number of single-stranded DNA products could be produced.
The team developed a novel looped bipedal DNA walking strategy. The two loops of dumbbell structured DNA probe contained DNAzyme sequences, which could not react with the track strands at the electrode interface initially (inactivated state).
When it was activated by the single-stranded DNA produced by the above strand displacement amplification, a looped structured DNA probe was formed from dumbbell probe. The bipedal walkers were activated for further interaction with the track strands at the electrode surface, inducing the changes of electrochemical response.
Based on the developed method, 2.2 aM (about 1.3 copy/μL) sensitivity could be realized under optimized experimental conditions, according to their results. “This method shows good selectivity,” said MIAO.
The clinical serum samples and throat swab samples were further tested and verified.
Through the analysis of abnormal electrochemical signals, the corresponding patients can be effectively identified from the healthy control groups, according to MIAO.
The strategy also provides a fast and sensitive new way for detecting DNA marker of acute infectious diseases.
This research was supported by the Science and Technology Cooperation Project between the Chinese and Australian Governments (Grant No. 2017YFE0132300). The research article, “DNA Hairpins and Dumbbell-Wheel Transitions Amplified Walking Nanomachine for Ultrasensitive Nucleic Acid Detection” has been published in ACS Central Science.

Figure 1. Illustration of the electrochemical sensing strategy based on looped bipedal DNA walking. (Image by MIAO’s group)
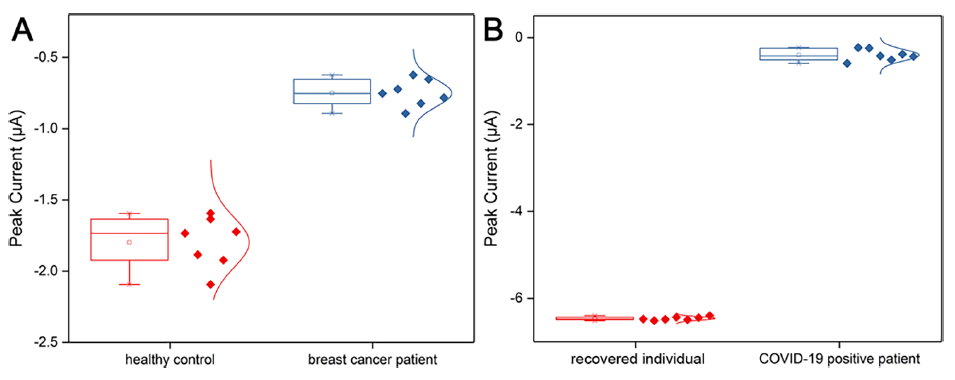
Figure 2. Box plots of SWV peak intensities for (A) breast cancer patients and healthy controls, (B) COVID-19 positive patients and recovered individuals. (Image by MIAO’s group)
Contact
XIAO Xintong
Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences (http://www.sibet.cas.cn/)
Phone: 86-512-69588013
E-mail: xiaoxt@sibet.ac.cn