Hybridization chain reaction is an enzyme-free nucleic acid polymerization reaction proposed by Dirks and Pierce in 2004. Target molecule triggers cascade hybridization reactions by several thermodynamically stable DNA fuel strands, producing ultra-long DNA nanostructures with nicks. Signal amplification of the target molecule can be achieved by the application of hybridization chain reaction.
Recently, a group of scientists in Suzhou Institute of Biomedical Engineering and Technology (SIBET) of the Chinese Academy of Sciences developed a new type of linear hybridization chain reaction, which couples magnetic nanoparticles and silver nanoparticles for highly sensitive and selective analysis of target nucleic acids.
Fe3O4 nanoparticles are firstly prepared for rapid magnetic separation and gold nanoparticles are formed on the surface for the immobilization of DNA strands. “Due to the huge specific area, a large number of DNA probe A can be immobilized,” said MIAO Peng, leading researcher of the study from SIBET.
This probe contains four functional regions, among which two are complementary to the target miRNA.
After introduction of DSN, DNA/RNA duplexes form and the digestion reactions release target miRNA strands for recycled reactions, and create single-stranded Probe B and C to further act on the DNA triangular prism structure at the electrode surface.
On the other hand, triangular prism structured three-dimensional DNA (TPDNA) is assembled at the electrode surface as the recognition layer. TPDNA shows geometrical rigid structure, which provides suitable sites to capture probes B and C.
The localized strands offer two single-strand regions, which are used to capture the fuel strands of H1, H2, H3 and H4. A long ladder-like DNA structure is formed which possesses multiple amino groups for immobilizing silver nanoparticles as strong electrochemical probes.
After measuring the silver stripping peak intensity, the initial target miRNA level can be evaluated. The nanomaterials involved in this work are fully characterized (Figure 1), and the corresponding reactions have been verified by polyacrylamide gel electrophoresis and electrochemical techniques (Figure 2). Under optimized experimental conditions, the detection limit of 5×10-18 mol/L can be achieved with good sequence selectivity.
Satisfactory results have been obtained in actual clinical sample assays, according to MIAO.
Results of the study was published in recent issue of Biosensors and Bioelectronics, entitled “Ultrasensitive miRNA biosensor amplified by ladder hybridization chain reaction on triangular prism structured DNA”.
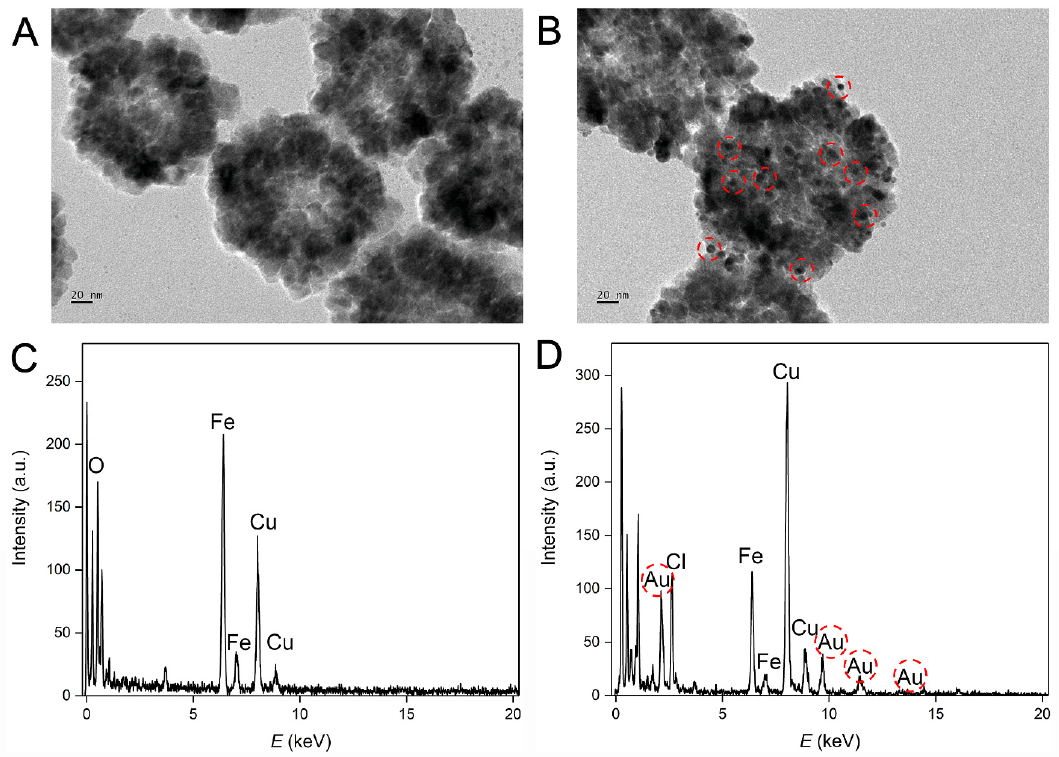
Figure 1. TEM images of (A) Fe3O4NPs and (B) Fe3O4NPs@AuNPs. EDS analysis of (C) Fe3O4NPs and (D) Fe3O4NPs@AuNPs. (Image by SIBET)
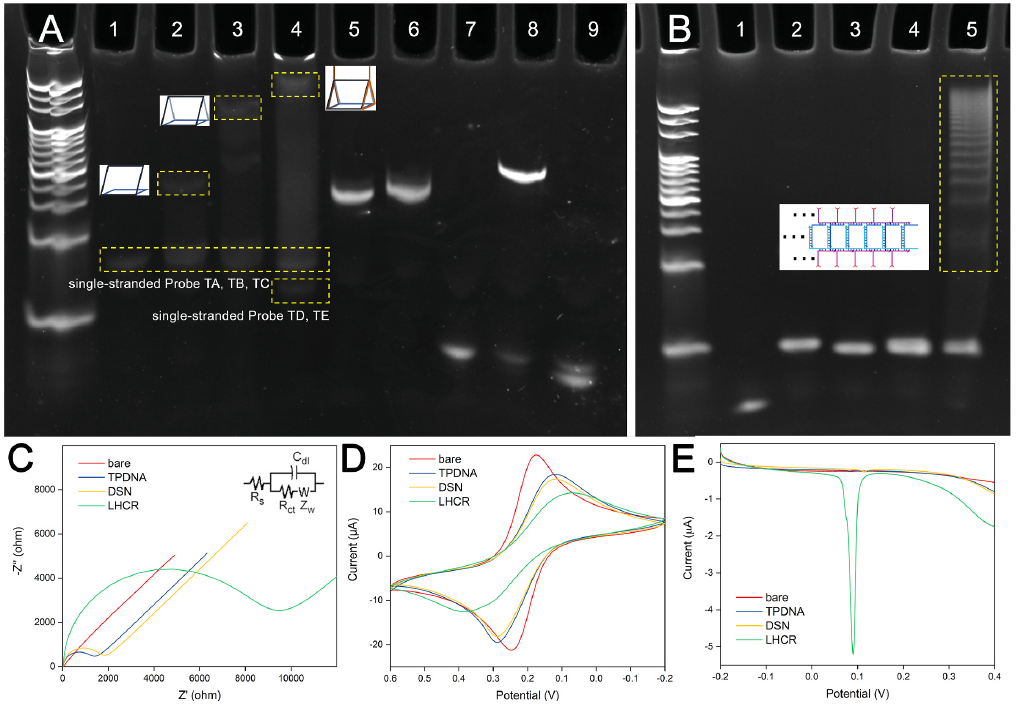
Figure 2. (A) PAGE analysis of TPDNA assembly and DSN catalyzed cleavage: 1, Probe TA; 2, Probe TA and TB; 3, Probe TA, TB, and TC; 4, Probe TA, TB, TC, TD and TE; 5, Probe A; 6, Probe A and DSN; 7, miR-141; 8, miR-141 and Probe A; 9, miR-141, Probe A and DSN. (B) PAGE analysis of LHCR: 1, Probe H3 and H4; 2, Probe H1 and H2; 3, Probe H1; 4, Probe H2; 5, Probe H1, H2, H3 and H4. (C) Nyquist diagrams, (D) cyclic voltammograms and (E) linear sweep voltammograms of bare electrode, TPDNA modified electrode, after DSN mediated reaction and then LHCR process. (Image by SIBET)
Contact
XIAO Xintong
Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences (http://www.sibet.cas.cn/)
Phone: 86-512-69588013
E-mail: xiaoxt@sibet.ac.cn