Messenger RNA (mRNA) is the molecule which carries genetic information from DNA and guides the synthesis of protein. It participates in many cellular cascades and exhibits substantial associations with certain diseases. The development of advanced approaches for mRNA sensing and imaging is of utmost importance for biological studies and early clinical diagnosis.
Researchers at the Suzhou Institute of Biomedical Engineering and Technology (SIBET) of the Chinese Academy of Sciences recently proposed an entropy-driven strand displacements strategy for highly sensitive determination and intracellular imaging of mRNA.
Entropy-driven strand displacement is a low-cost and highly efficient isothermal amplification approach, which does not require any actions of polymerase and nuclease. It provides toolboxes to implement DNA computing, and assists signal amplification with target-triggered multiple cycles.
This newly-developed enzyme-free fluorescent biosensor is constructed based on entropy-driven strand displacements around three-dimensional DNA nanostructures. It can achieve high sensitive detection of target mRNA, and provide an intracellular bioimaging protocol.
Tetrahedral DNA with four pendant linear sequences provides excellent molecular scaffold loading at multiple sites for target mRNA-initiated cascade toehold strand displacement reactions.
In addition, a duplex is formed by three single-stranded DNAs for initial fluorescence signal quenching and target mRNA recognition. After target mRNA-initiated strand displacement cycles around TDNA, the fluorescence resonance energy transfer state of the probe is shifted from “off” to “on”. The recovered fluorescence response indicates the level and position of target mRNA inside cells (Figure 1).
The feasibility of reactions has been confirmed by fluorescence and electrochemical techniques (Figure 2). Under optimized experimental conditions, the detection limit of 3.3x10-16 mol/L can achieve good sequence selectivity.
This method can precisely probe the information of intracellular mRNA, which holds intriguing potential for biological and clinical applications, according to the researchers.
Result of the study entitled “Entropy-Driven Strand Displacements Around DNA Tetrahedron for Sensitive Detection and Intracellular Imaging of mRNA” was published in the recent issue of Small Structures.
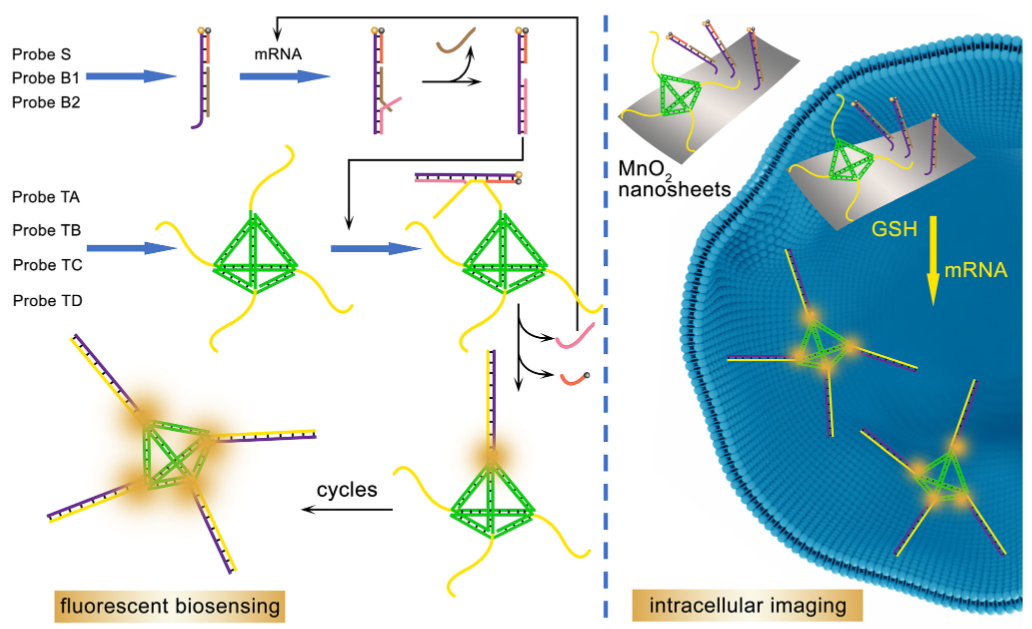
Figure 1. Illustration of the entropy-driven strand displacements around TDNA for mRNA assay. (Image by SIBET)
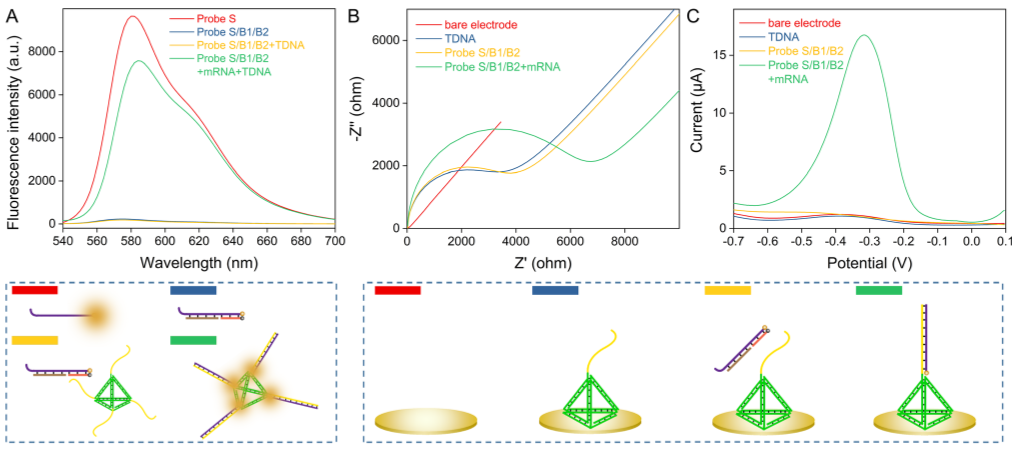
Figure 2. Fluorescence and electrochemical characterization of entropy-driven chain displacement reactions. (Image by SIBET)
Contact
XIAO Xintong
Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences (http://www.sibet.cas.cn/)
Phone: 86-512-69588013
E-mail: xiaoxt@sibet.ac.cn